By Josh Sanes
For many years, our lab has used the retina, a particularly accessible part of the brain, to find out how neurons form the specific connections that lead to assembly of complex information-processing circuits. One impediment, not unique to retina, has been lack of a “parts list” of its cell types. A way out of this impasse came with the invention of high-throughput single cell RNA sequencing (scRNA-seq), which maps gene expression profiles of many thousands of cells quickly and cheaply enough to enable comprehensive identification of essentially all cell types in a tissue. We were lucky enough to collaborate with the inventors of one such method, called Drop-seq, as it was being perfected. We were so awestruck by the initial results that we went on to generate a complete cell atlas of the mouse retina, identifying some 130 neuronal types and over a dozen non-neuronal types over the course of four studies (2016, 2019, 2020, 2023). (We then used similar methods to generate atlases of human, macaque, chick and zebrafish retina.
This brings us to the recent paper by Hahn et al. We wondered if we could generate retinal cell atlases from many species, and use them to ask which cell types are conserved, which are present in only some species, and so on. We thought the answers could help us study the evolution of cell types, just as many others have studied the evolution and conservation of genes. In addition, we were especially interested in a type of retinal ganglion cell (RGC) called the midget, which comprises nearly 90% of all RGCs in humans. RGCs are the sole conduit of visual information from our eye to our brain, so their function is critical for vision, and their dysfunction leads to blindness. Yet, no one had found non-primate orthologues of midgets, and they were generally thought to be a primate innovation. We hoped that by comparing enough species, we might be able to fill in the blanks and find some orthology.
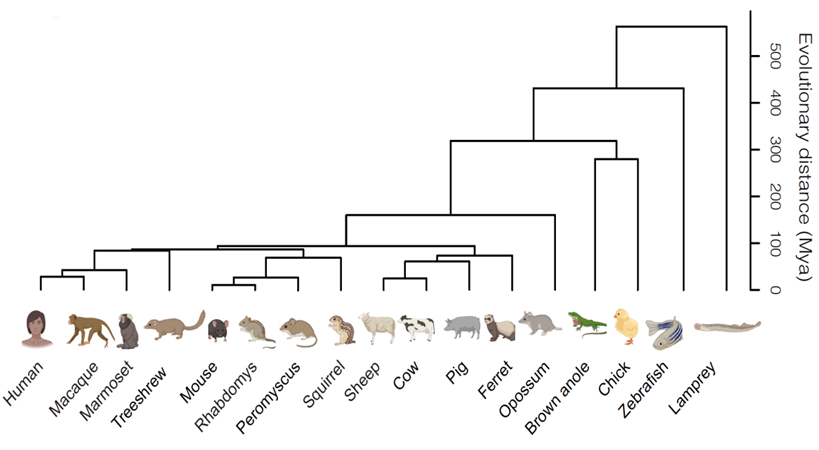
Hahn et al found striking conservation of cell types among retinas of these 17 species, whose last common ancestor lived over 500 million years ago.
The project began with two postdoctoral fellows, Aboozar Monaverfeshani and Karthik Shekhar; it continued as a collaboration with Karthik after he became a faculty member at UC Berkeley and recruited his first graduate student, Josh Hahn, to the team. We ended up generating retinal cell atlases from 17 species. The comparisons showed extraordinary conservation of cell types – and the result has become more impressive as we’ve since added additional cells and species. Most satisfying was that we did find midget orthologues. Surprisingly, in mouse, they are a very large and minor type, the opposite of their primate versions. A next step will be to search for the changes in gene expression that underlie their increased abundance and decreased size during primate evolution. More generally, study of these mouse orthologues may help us understand more about the developmental programs that generate midget RGCs, and the ways in which they are affected by injury and disease.
Josh Sanes is the Jeff C. Tarr Professor of Molecular and Cellular Biology at Harvard and the Founding Director of the Center for Brain Science.
We are highlighting papers from Harvard labs published recently in Nature as part of of the NIH’s Brain Research Through Advancing Innovative Neurotechnologies initiative Cell Census Network (BICCN). Learn more about this initiative here.
Learn more in the original research article:
Evolution of neuronal cell classes and types in the vertebrate retina.
Hahn J, Monavarfeshani A, Qiao M, Kao AH, Kölsch Y, Kumar A, Kunze VP, Rasys AM, Richardson R, Wekselblatt JB, Baier H, Lucas RJ, Li W, Meister M, Trachtenberg JT, Yan W, Peng YR, Sanes JR, Shekhar K.Nature. 2023 Dec;624(7991):415-424. Epub 2023 Dec 13.
News Types: Community Stories
