By Wenjun Yan
Glaucoma is the leading cause of irreversible blindness worldwide. It results from the death of retina ganglion cells (RGCs) that carry visual information from the eye to the rest of the brain. The major modifiable risk factor for glaucoma is an increased intraocular pressure (IOP), which is generated by excessive production or reduced drainage of aqueous humor from the anterior chamber of the eye through tissues in the anterior segment (see Figure). Advancing therapeutics for glaucoma depends on improved understanding of (1) the tissues that regulate IOP and (2) the RGCs that fall victim to it. To address these questions, we recently used high throughput single cell RNA sequencing (scRNA-seq) to classify and characterize cell types in the anterior segment and the retina of the human eye.
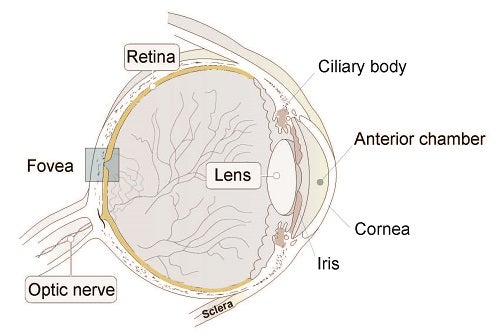
Schematic of the human eye.
Our findings have come out in three new studies that we highlight here. In the first study, we built a human cell atlas covering the two pathways through which the aqueous humor is drained. One is called the conventional outflow pathway, in which aqueous humor exits through a structure called the trabecular meshwork. A second pathway, called uveoscleral, provides an alternate exit strategy. Most current drugs used to treat glaucoma target one or both of these pathways, improving outflow and thereby decreasing intraocular pressure.
We identified 19 cell types in these tissues with distinct molecular markers, known or novel. We validated many of them in tissue with in situ hybridization or immunostaining. This is to our knowledge the first transcriptomic cell atlas of human trabecular meshwork and the surrounding tissues. It provides access to molecular identification and genetic manipulation of these tissues (van Zyl et al., 2020).
Next, we turned to the retina. Although we and others had studied RGCs in rodent retina in details, human retina differs in a critically important and clinically relevant respect. Human and most primates have high acuity vision thanks to a small central area in the retina called fovea. It occupies only ~1% of the retina surface, while provides ~50% of the input to primary visual cortex. If the fovea is lost but the rest of the retina is intact, people are legally blind. To provide insight into this specialization, we utilized scRNA-seq to profile retina cells from human fovea and peripheral retina separately. The human retina has a complicated cellular composition with in total 58 cell types from 6 classes: photoreceptor, horizontal, bipolar, amacrine, retinal ganglion and non-neuronal cells. Nearly all types are shared between fovea and peripheral retina, with notable differences in gene expression and type proportion between the fovea and peripheral cohorts (Yan et al., 2020a).
Next, we used these data to map the expression of genes implicated in glaucoma either as causal genes with Mendelian inheritance or as susceptibility loci identified in genome-wide association studies. While many genes associated with high IOP were preferentially expressed in the anterior segment, others that suggested to be IOP-independent glaucoma risk genes were more predominantly in the retina.
Finally, we compared human cell types and gene expression profiles with those from model organisms, which are used for both basic and translational research. For retina, we used data sets that we had previously collected from mouse (Shekhar et al., 2016; Tran et al., 2019; and our latest round in Yan et al., 2020b) and Macaque (Peng et al., 2019). For anterior segment, we profiled tissues from four animal models frequently used in the study of aqueous humor outflow pathways and glaucoma——Macaca fascicularis (crab-eating macaque), Macaca mulatta (rhesus macaque), Sus scrofa (pig), and Mus musculus (mouse). We found that while broad cell classes were generally conserved across all above species, some types within class were not. As expected, human types were more similar to those of macaques than to those of other mammals.
Together, these results provide a foundation for future study of glaucoma and other retinal diseases, and help guide tests of therapeutic strategies in these models.
Wenjun Yan is a postdoctoral fellow in the lab of Josh Sanes, in the Department of Molecular and Cellular Biology at Harvard.
Learn more in the original research articles:
van Zyl, T. et al. Cell atlas of aqueous humor outflow pathways in eyes of humans and four model species provides insight into glaucoma pathogenesis. Proc Natl Acad Sci U S A 117, 10339-10349, doi:10.1073/pnas.2001250117 (2020).
Yan, W. et al. Cell Atlas of The Human Fovea and Peripheral Retina. Sci Rep 10, 9802, doi:10.1038/s41598-020-66092-9 (2020).
Yan, W. et al. Mouse Retinal Cell Atlas: Molecular Identification of over Sixty Amacrine Cell Types. J Neurosci 40, 5177-5195, doi:10.1523/JNEUROSCI.0471-20.2020 (2020).
Additional References:
Shekhar, K. et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308-1323 e1330, doi:10.1016/j.cell.2016.07.054 (2016).
Tran, N. M. et al. Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron 104, 1039-1055 e1012, doi:10.1016/j.neuron.2019.11.006 (2019).
Peng, Y. R. et al. Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222-1237 e1222, doi:10.1016/j.cell.2019.01.004 (2019).
News Types: Community Stories
