By Connie Cepko
As March opened with the SARS-CoV-2 pandemic upon us, Brian Rabe, a graduate student in the Cepko Lab at HMS in the Department of Genetics and HHMI, began to consider how he could help. Brian had been working with nucleic acids since he was in high school, and is deeply knowledgeable about the many ways in which one can detect RNA or DNA. He recognized that the standard diagnostic method for detection of the genome of SARS-CoV-2, the reverse-transcription polymerase chain reaction (RT/PCR) method, was limited in terms of cost and scale. Some of the machines used for RT/PCR in clinical settings cost a couple hundred thousand dollars, purification methods were in short supply and cartridges for the machines were in short supply. To bypass these problems, Brian turned to the reverse-transcription loop-mediated isothermal amplification (RT/LAMP) method. This method also uses a nucleic acid amplification reaction, much like RT/PCR, and thereby gives great sensitivity and specificity. However, rather than rely upon expensive instruments and limited materials, the RT/LAMP method can simply be run by placing reagents in a tube at 65°C. Even a water bath in a kitchen, set up using a sous vide, can do the job. The reaction is capable of generating microgram quantities of viral DNA in 30 minutes at a single reaction temperature, with results read out by a color change. The method had been developed over 20 years previously and was deployed in field situations for detection of plant or animal pathogens. One innovation on the method developed by Rabe and Cepko was to incorporate another old, but simple, protocol for purifying the viral RNA from biological samples. The method calls for the addition of glass powder to the sample, which binds the viral RNA, allowing it to be isolated away from contaminating materials, thereby improving sensitivity. Glass milk has no supply chain limitations; it is the same material used in the ceramics industry. The estimated cost of this step is 7 cents per sample. This method has been ready to scale and distribute since April but lack of approval and leadership from the Federal government has hampered commercial development. New leadership will hopefully change this situation in the near future, as we are still in desperate need for more diagnostics.
You can learn more about the reaction here: https://www.youtube.com/watch?v=L5zi2P4lggw
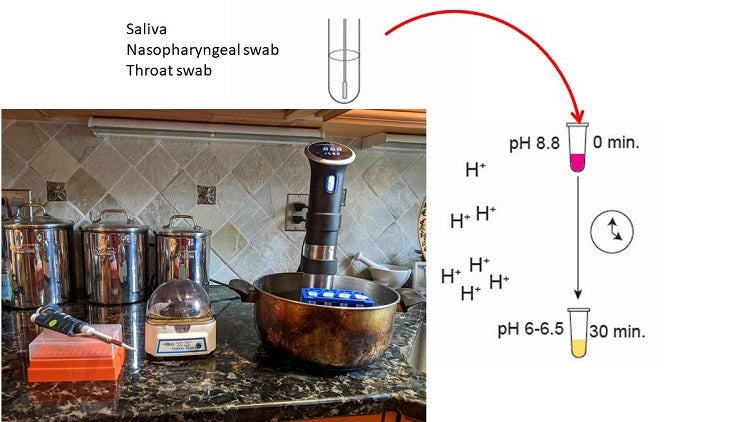
The RT/LAMP reaction can be run with minimal equipment and expertise. A variety of sample types can first be heat-treated to inactivate the virus, then placed in a reaction tube with reagents for the RT/LAMP reaction, and incubated in a simple water bath for 30’. As DNA amplification occurs, protons are produced, leading to a color change from pink to yellow. The setup shown here is not high throughput, but the method can be scaled using robotics. Pooling of samples for surveillance testing, e.g. of classrooms, is also being developed by Randy True of FloodLAMP in San Francisco.
Connie Cepko is a Professor of Genetics and Ophthalmology at Harvard Medical School and an Investigator at the Howard Hughes Medical Institute.
Learn more in the original research article:
Rabe, B. A., & Cepko, C. (2020). SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proceedings of the National Academy of Sciences of the United States of America, 117(39), 24450–24458. https://doi.org/10.1073/pnas.2011221117
News Types: Community Stories
