By Xin Jin
Approximately 1 in every 5 adults in the United States, that is 47.6 million people, experience some form of mental illness in their lives. Through modern genetics, we now know that our vulnerability to many psychiatric disorders is highly genetic. They are largely heritable: zygotic twin studies have estimated heritability for bipolar disorder, schizophrenia, and Autism Spectrum Disorders (ASD) to be 50-90%, which is much higher than that of many other diseases such as breast cancer.
Thanks to the recent advances and heroic efforts in human genetics, hundreds of risk genes have been identified by whole exome sequencing of patients with ASD and intellectual disability. Systemic, causative genetic studies of these hundreds of ASD risk genes are urgently needed but generating knockout animal models for individual genes is time-consuming and costly. Additionally, the brain is a complex organ comprised of many cell types, including excitatory neurons, inhibitory neurons, and glia cells such as oligodendrocytes. Although ASD has a developmental origin, it is not known whether different risk variants can affect the same cell types and/or molecular networks.
To begin to address some of these questions, we turned to Perturb-Seq. Perturb-Seq is a platform developed by the labs of Aviv Regev, Jonathan Weissman and Ido Amit in 2016, which combines the pooled gene editing–specifically, using CRISPR-Cas9 to introduce mutations in specific loci in the genome—with single-cell RNA sequencing to readout the genetic perturbation effect (see Dixit et al., Adamson et al., Jaitin et al. Cell 2016). It has been used in vitro to study various questions such as immune activation in dendritic cells.
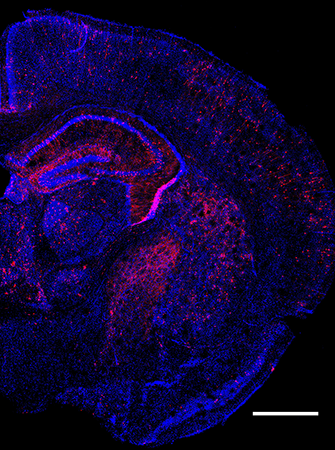
Coronal image of the mouse brain at P7 with in vivo Perturb-seq (perturbed cells in red).
We sought to expand this approach to in vivo, to build a system to study the functions of diverse ASD risk gene mutations in the context of a developing brain in utero in living animals. Using the CRISPR-Cas9 genome editing technique, we introduced frameshift mutations in 35 ASD risk genes in pools, within the developing mouse neocortex in the embryos, followed by single-cell transcriptomic analysis of perturbed cells from the early postnatal brain. We found that perturbations in 9 different risk genes significantly affected 4 different cell classes (cortical excitatory neurons, inhibitory neurons, astrocytes, and oligodendrocytes). Working closely with computational biologist Sean Simmons, we used machine learning-based approaches and gene network analysis and unveiled 5 gene networks with significantly altered expression levels, which represent key cellular effects by cohort of risk gene perturbations.
These widespread effects highlight the diverse landscape and cell-type specificity of ASD. We validated several results from this screen and conducted a number of follow-up studies to identify the molecular networks affected by ASD risk gene perturbations. We also compared my results with human ASD postmortem brain data and uncovered some shared effects in both mouse and human, such as decreases in the peptide somatostatin (SST) in interneurons and the neuronal protein neuritin 1 (NRN1) in excitatory neurons.
Together, our findings suggest that ASD is a disorder that affects diverse cell types including both neurons and glia, and that Perturb-seq uncovers shared effects that are observed across species, representing key cellular effects related to these risk loci. In vivo Perturb-seq is a scalable tool for functional studies of large gene panels in complex tissues and diverse organisms. In the future, in vivo Perturb-seq can be coupled with other readout methods such as spatial readout of RNA and protein. This will enable us to better define both cell-autonomous and non-autonomous/network effects by the disease risk genes. This approach represents a new area of neurobiology—by connecting genomic technology development with rigorous dissection of molecular mechanisms, we hope to gain new insights about how complex inputs are integrated in the developing brain.
Xin Jin is a Junior Fellow at the Harvard Society of Fellows, working with Feng Zhang (McGovern Institute for Brain Research at MIT and Broad Institute) and Paola Arlotta (Department of Stem Cell and Regenerative Biology at Harvard).
Learn more in the original research article:
In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Jin X, Simmons SK, Guo A, Shetty AS, Ko M, Nguyen L, Jokhi V, Robinson E, Oyler P, Curry N, Deangeli G, Lodato S, Levin JZ, Regev A, Zhang F, Arlotta P. Science. 2020 Nov 27;370(6520):eaaz6063. doi: 10.1126/science.aaz6063.
News Types: Community Stories
