By Meng Zhang
A cell atlas of the brain, an extremely complex machine, catalogs the brain’s fundamental units—the cells, with their properties and locations—and also illustrates how they are connected to form functional circuits. Recent advances in technology have shed light on the road towards a comprehensive brain atlas: Single-cell sequencing methods systematically identify cell types and quantify the heterogeneity of tissues. Spatially resolved transcriptomics elucidate cellular heterogeneity while retaining spatial information in intact tissues, which is crucial in gaining a systematic understanding of how the brain works. Here, using a single-cell transcriptome-imaging method, multiplexed error-robust fluorescence in situ hybridization (MERFISH), we sought to build a molecularly defined, spatially-resolved and projection-annotated cell atlas of a brain region in the mouse that controls movement, the primary motor cortex (MOp).
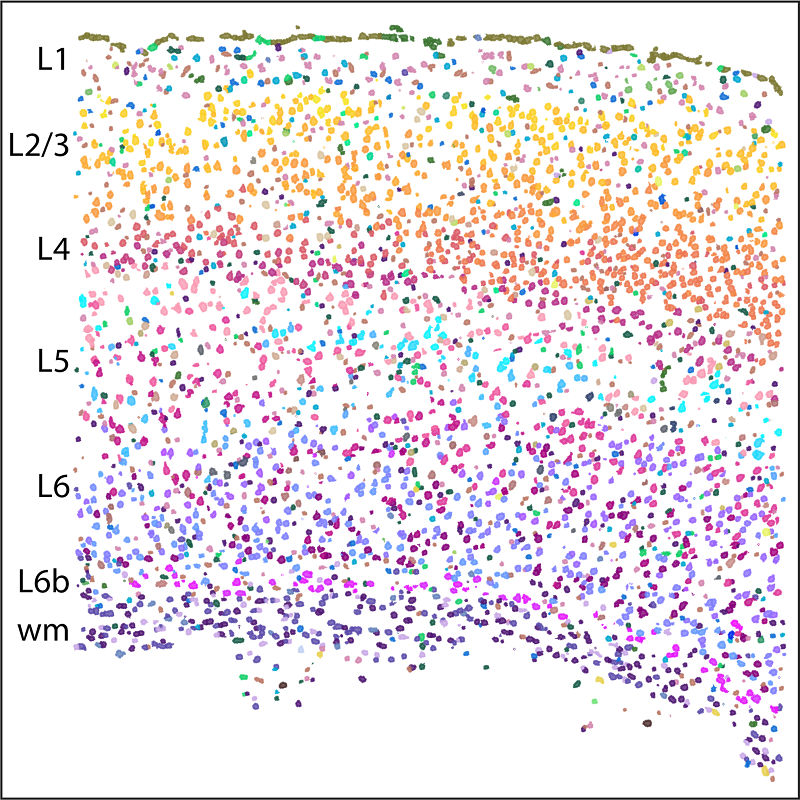
MERFISH, a single cell transcriptome imaging method, identifies 95 molecularly defined cell types in the mouse primary motor cortex (MOp) and records their spatial information in intact tissue. These cells form complex spatial organizations with an overall layered structure as shown here in one of the coronal slices imaged by MERFISH. L1 to L6 are layers of cortex; wm refers to white matter (myelinated axons).
We profiled ~300,000 cells in MOp and identified 95 neuronal and non-neuronal cell types, which showed excellent correspondence with those defined by single-cell sequencing methods. The cell types in MOp formed highly complex spatial organizations with an overall laminar appearance dominated by the densely packed layers of the intratelencephalic (IT) neurons. Surprisingly, most inhibitory neuronal clusters also adopted laminar organizations, with many residing in one cortical layer, some in two. We observed both discrete and continuous changes between different cell types in both molecular and spatial profiles and found that their distinctness in molecular and real spaces are often correlated. One striking example is the largest neuronal population in MOp, the IT neurons. The 19 IT cell clusters determined by MERFISH formed a largely continuous gradient across the cortical depth axis, with highly correlated gene expression profiles and cortical depths of individual cells.
We further integrated MERFISH with retrograde tracing to map cell types and their projection properties simultaneously at a single cell level. Our proof-of-principle measurements revealed a complex multiple-to-multiple projection network between MOp and three other brain regions that were injected. As an imaging-based technique, we envision that MERFISH can be combined with various other techniques to align spatially resolved transcriptomics with different functional properties of the cells in the brain and other tissues.
This work is supported by the NIH’s Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative Cell Census Network (BICCN), which aims to identify and catalog the diverse cell types in human, monkey and mouse brain, and is published on Nature as a part of BICCN’s first major installment, a multimodal cell census and atlas of mammalian primary motor cortex. See the complete BICCN consortium package here: https://www.nature.com/collections/cicghheddj
Meng Zhang a postdoctoral fellow in Prof. Xiaowei Zhuang’s lab in the Department of Chemistry and Chemical Biology at Harvard University.
Learn more in the original research article:
Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Zhang M, Eichhorn SW, Zingg B, Yao Z, Cotter K, Zeng H, Dong H, Zhuang X. Nature. 2021 Oct;598(7879):137-143.
News Types: Community Stories
